Self-Assembly in Membranes and Molecular Dynamics on Surfaces
Novel carbon microelectrodes enable in vivo detection of neurotransmitters. However, atomistic detail on the diffusion and orientation of neurotransmitters on these surfaces is lacking. In collaboration with Prof. B. Jill Venton, we employed molecular dynamics simulations to investigate the surface structure and diffusion of dopamine (DA) on the pristine basal plane of flat graphene, finding that all DA species rapidly adsorb to the surface and remain adsorbed, even without a holding potential or graphene surface defects. We also found that solvation has a large effect on DA surface dynamics on both flat graphene (Jia, ChemPhysChem, 2022) and curved carbon nanotube surfaces (Jia, Molecules, 2022), which results in diffusivities that depend on adsorbate charge, oxidation state, and surface curvature. Our simulations also show that diffusion differs significantly on H-terminated CNTs and graphene ribbons, and simulations of DA on the edge plane of stacked graphenes are underway.
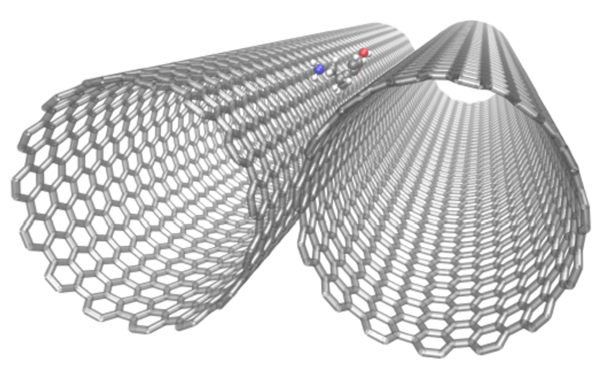
Figure 1: Simulated Carbon Nanotubes with Adsorbed Dopamine. Dopamine diffuses into and remains in the exterior groove formed by two solvated and parallel (15,15)-carbon nanotubes, enhancing diffusivity along the carbon nanotube axis. The solvating water molecules were omitted for visual clarity (Jia, Molecules, 2022).