Genetic Analysis
We are developing a fully-integrated microdevice capable of DNA extraction, PCR amplification and the subsequent electrophoretic separation specifically for the detection of T-cell lymphoma (TCL). While we have recently reported a fully-integrated device for the detection of bacteria [1], the interrogation of human genomic DNA from whole blood provides new challenges, especially when detecting gene rearrangements correlative with cancer. Detection of TCL involves the PCR amplification of select sequences in the T-cell receptor gene that are likely to have undergone gene rearrangement. These PCR fragments represent a polyclonal cell population in normal individuals and a monoclonal cell population in patients with lymphoma in a way that can be discriminated by electrophoretic separation. In clinical labs, the PCR that follows DNA extraction requires ~3 hours followed by a 40 min capillary electrophoresis separation under single-stranded conditions and utilizing 4-color detection. While integration of processing steps is a large advantage over traditional methods, the reduction in the times associated with these lengthy processes is also an important benefit.
1. Easley, C.J., Karlinsey, J.M., Bienvenue, J.M., Legendre, L.A., Roper, M.G., Feldman, S.H., Hughes, M.A., Merkel, T.J., Ferrance, J.P., Landers, J.P. PNAS, 2006, 109(51), 19272-19277.
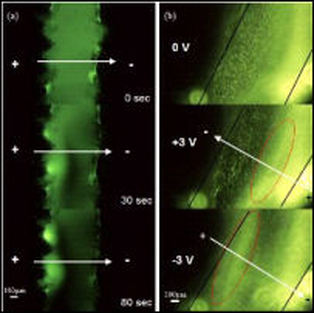
In many separation scenarios, a simple preconcentration step between purification steps is highly desired. For example, the elution step on a microchip solid phase extraction column dilutes the DNA and raises the threshold for downstream amplifications. To solve this problem, we are developing electric field-flow based glass/PDMS microdevices to recover concentrated DNA samples from upstream extraction/purification steps.
Laser-induced fluorescence (LIF) is the most common form of detection on microchips. Typically, a confocal setup is used, where incoming laser light is focused into a microchannel through an objective, and the fluorescence emission is collected and sent to a photomultiplier tube (PMT) for detection. Increasing the number of fluorophores to be detected in the sample creates a need for separating out different emission lines. We are using an acousto-optic tunable filter (AOTF) to select these emission lines. The AOTF acts as an electronically tunable spectral bandpass filter. It is made up of a single optically-active crystal bonded to a piezoelectric. By applying different radio frequencies to the piezoelectric, acoustic waves are propagated through the crystal, and a single wavelength of light can be separated from a multi-color source. This allows for a separation of multiple species spectrally, even without temporal resolution. We plan on using this device to perform multicolor detection of tagged DNA bases for clinical and forensic applications.